100-plex Grant Program—PhenoCycler-Fusion
Deep Spatial Phenotyping for “Hallmarks of Cancer”
Grant Program Awardee

Resident Physician
Harvard Radiation Oncology Program
Massachusetts General Hospital
Dr. Ryan Park received his MD from Harvard Medical School and is currently a resident physician in the Hwang Lab at the Center for Systems Biology at Massachusetts General Hospital. Dr. Park’s research focus is in immunology and systems biology. You can review a list of his recent publications in his Harvard Catalyst Profile.
Study Proposal
Pancreatic ductal adenocarcinoma (PDAC) has exceptionally poor prognosis and is entirely resistant to current immunotherapies. PDAC has historically been split into 1) classical-like and 2) basal-like consensus subtypes, with the latter associated with worse survival. We recently refined this taxonomy (Hwang et al Nature Genetics in press) by developing a robust single-nucleus RNA-seq (snRNA-seq) approach to study a cohort of resected primary tumors and identified novel PDAC lineage subtypes associated with treatment-resistance and poor prognosis, including a “neural-like progenitor” (NRP) subtype that expresses genes normally found only in the brain. By using digital spatial profiling (NanoString GeoMx) on the resected tumors, we found that CD8 T cells are enriched in regions of tumor with NRP differentiation, which is surprising given that high CD8 T cell infiltration is generally associated with improved prognosis.
We propose to characterize the immune landscapes associated with each of the distinct PDAC malignant subtypes identified by our recent work. By applying the 100-plex Akoya panel to our resected primary PDAC tumors, we would identify the phenotype, cell state, and functionality of lymphoid and myeloid cells. We would use RNAscope on a consecutive section to classify the malignant cell subtypes based on transcription factor expression. In preliminary analysis of our GeoMx and snRNA-seq data, we have observed marked differences in the immune landscapes associated with each PDAC malignant subtype, including differences in expression levels of beta-2-microglobulin and HLA, cytokines, and genes associated with CD8 T cell effector function, differentiation, and exhaustion.
The Akoya platform would give us the opportunity to extend these spatial studies to the single-cell and protein level in order to deeply characterize the intra-tumoral heterogeneity in the tumor immune microenvironment of PDAC, revealing potentially distinct mechanisms of immune evasion that can inform combinatorial therapeutic strategies and thereby advance precision oncology for this lethal disease.
Deep Spatial Phenotyping for “Hallmarks of Cancer”
Akoya Biosciences invites scientists involved in oncology research to apply for a deep spatial phenotyping “Hallmarks of Cancer” grant award.
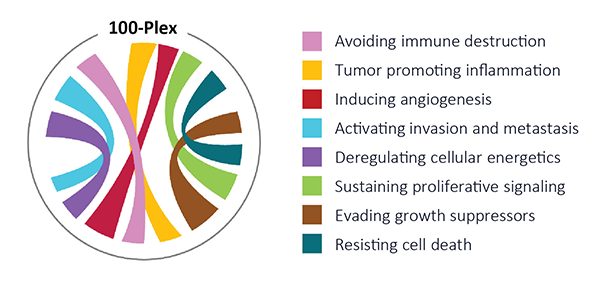
The PhenoCycler™-Fusion System delivers unprecedented speed and depth enabling researchers to scale up unbiased discovery. Combined with the 100-plex “Hallmarks of Cancer” panel, this assay provides deep insights into the eight functional pathways of cancer.1
The grant recipient will receive:
- Deep spatial phenotyping data to reveal the presence of 100 cancer biomarkers
- Spatial insights for up to 3 FFPE tissue samples
- An assay report on results of the PhenoCycler-Fusion workflow summarized by Akoya’s application team
Apply Now for the 100-plex Grant Program—
PhenoCycler-Fusion System
The grant program is subject to the 100-plex Grant Program—PhenoCycler-Fusion Terms and Conditions which contain eligibility restrictions. No purchase is necessary to enter; void where prohibited.
References
1. Hanahan D (2022) Hallmarks of Cancer: New Dimensions. Cancer Discov 2022, 12:31-46.