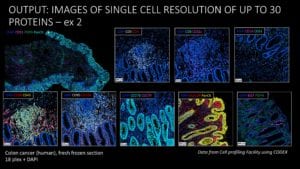
In our recent webinar, we heard from speakers at the Fred Hutchinson Cancer Research Center, who discuss how they successfully implemented and utilize the Vectra® Polaris™ and Vectra® 3 automated, multiplexed tissue imaging platforms in their core lab.
Our first speaker was Dr. Savanh Chanthaphavong, the Director of the Experimental Histopathology Core Lab at the Fred Hutchinson Cancer Research Center. The Experimental Histopathology Core Lab provides comprehensive histology and immunohistochemistry services, including cell and tissue harvesting, cell pellet-making, TMA construction, tissue processing, and H&E/special staining. They also perform routine IHC and antibody optimization and screening.
Dr. Chanthaphavong explained how the use of serial sections and chromogenic immunohistochemistry (IHC) is insufficient to study the tumor microenvironment.
“Using serial sections and chromogenic IHC has been difficult to study the dynamic and phenotypic changes of cell subsets during activation or differentiation, or to really understand how cell subsets respond to treatment,” said Dr. Chanthaphavong.
To overcome these challenges, Dr. Chanthaphavong and her team made the decision to adopt Akoya’s Vectra® 3 and Vectra® Polaris™ imaging systems for multiplexed, high-throughput spatial analysis of FFPE tumor samples. Investigators can analyze up to eight tissue biomarkers simultaneously on the Vectra® Polaris™ system, and up to six on the Vectra® 3. By adopting these platforms, the Experimental Histopathology Core Lab was able to increase their output and turnaround time to better support Fred Hutchinson researchers.

The Vectra® Polaris™ Automated Quantitative Pathology Imaging System
“We’ve been quite successful with our multiplex service, and the Vectra® imaging system has helped our investigators to perform cellular phenotyping in the tumor microenvironment,” said Dr. Chanthaphavong.
Our next speaker was Dr. Kristin G. Anderson, a Senior Postdoctoral Research Fellow in the Greenberg Lab at Fred Hutchinson. She offered her perspective as a frequent user of the multiplex immunohistochemistry (mIHC) services from the Experimental Histopathology Core Lab.
One of the main hypotheses being tested in the Greenberg lab is how engineered T cells can be used to treat ovarian cancer, specifically high-grade serous carcinoma – the most common and most deadly ovarian cancer.
Dr. Anderson and her colleagues have developed CD8 T cells engineered to express a high-affinity T-cell receptor specific for an ovarian cancer antigen. They aim to use these T cells to slow tumor growth and prolong survival in patients.
The team uses the ID8VEGF mouse model in their research because it recapitulates characteristics of human ovarian cancer. Vascular endothelial growth factor (VEGF) and mesothelin, a self-protein thought to promote excessive growth, are both overexpressed in this model as well as in high-grade serous ovarian cancer.
In Dr. Anderson’s presentation, she focused on her lab’s efforts to target mesothelin with their engineered T cells. In an early experiment, they observed that while engineered mesothelin-specific T cells can infiltrate ID8VEGF tumors, they do not persist for very long. Additionally, when the engineered T cells were taken out of the tumor 21 days after infiltration, it was observed that the cells lost their ability to make anti-tumor cytokines. This correlated with the expression of markers thought to be inhibitory receptors.
To understand this phenomenon, the Greenberg lab worked closely with the Experimental Histopathology Core Lab to characterize the tumor microenvironment and determine how the expression of suppressive ligands might be contributing to the dysfunction of T cells in tumors.
Dr. Anderson mentioned a paper published this year, which evaluated the expression of inhibitory receptors on T cells from human ovarian cancer using flow cytometry. Using this technique, they quantified the number of inhibitory receptors on T cells extracted from tumors, showing that T cells can display between 0 to 3 inhibitory receptors.
She noted that while this paper was important, it was lacking in spatial context.
“This [paper] suggests that there are multiple negative signals that these T cells are getting in the tumor microenvironment. But one thing that this particular analysis was lacking, because it was a flow cytometry experiment, was spatial resolution,” said Dr. Anderson.
Using mIHC services from the Experimental Histopathology Core Lab, Dr. Anderson’s team imaged tumor tissues to identify where these inhibitory receptors are being expressed in the ID8VEGF mouse model. The Core Lab developed panels for the Vectra® system to visualize the engineered T cells and inhibitory receptors Lag-3, PD-1, and Tim-3. They observed that the T cells with inhibitory receptors were clustering in the center of the tumor, suggesting that there is a spatial component involved in the cells recognizing antigens.
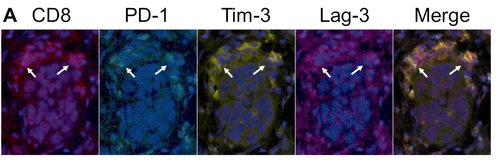

CD8+ T cells in human (top) and mouse (bottom) ovarian tumors can
co-express PD-1, Tim-3 and Lag-3 (Images courtesy of Dr. Kristin Anderson)
From the results of their investigation, Dr. Anderson’s team decided to administer repeat doses of CD8 T cells engineered to target ovarian cancer antigens to determine if this would negate T cell dysfunction following the initial dose. They found that repeat doses, in combination with chemotherapy and a vaccine boost to promote T cell persistence, significantly prolonged survival in the mouse model.
Dr. Anderson emphasized the importance of spatial context in yielding further insights into T cell function.
“Spatial resolution and multiplexed IHC imaging will allow us to understand some of the mechanisms driving T cell dysfunction,” she said.
We’re grateful to Dr. Chanthaphavong and Dr. Anderson for sharing their experiences implementing the Vectra® system and in using it as a core lab service. In case you missed the webinar, you can watch the recording here.