Dr. Charlotte Stadler is the head of the Cell Profiling Facility at SciLifeLab, a Swedish national resource center which offers life science researchers access to cutting-edge technologies and the expertise of internationally-recognized scientists. The Cell Profiling Facility is closely intertwined with the research lab of Dr. Emma Lundberg, who leads the Cell Atlas project, part of the Human Protein Atlas.
In 2020, the facility began offering highly multiplexed immunofluorescence services using the CODEX platform. We spoke to Dr. Stadler to learn about the types of CODEX projects they’ve been working on and how their services will evolve in the future.
Tell us about SciLifeLab. What kinds of services do you offer?
SciLifeLab is a Swedish national resource center for life sciences that started in 2010. We have a very broad portfolio of infrastructure services via our facilities, that are closely interwined with our community of researchers in areas such as biomedicine, ecology and evolution. We offer many services within genomics, proteomics, metabolomics, advanced light microscopy, and chemical biology to mention a few technology areas. I am heading a facility called Cell Profiling, that is focused on imaging using immunofluorescence. Last year we initiated service also in highly multiplexed imaging using the CODEX platform.
How is your team at the Cell Profiling Facility organized?
One of the strengths with SciLifeLab is that the infrastructure, the facilities that provide service, are integrated into the research groups. In our case, we work closely with professor Emma Lundberg, who is also the Director of the Cell Atlas, part of the Human Protein Atlas (www.proteinatlas.org).
This means that we are sharing the labs and many of the instruments that we use. And we also have a high degree of overlapping knowledge between the facility staff and members of the research team. This makes it easy to collaborate and share also personal resources if needed. Since our facility is still rather small, this makes us less vulnerable. But the biggest advantage is of course the closeness to the research conducted in Emma´s lab and that we can work together and implement new protocols, conjugated antibodies, etc. to remain at the forefront and offer competitive service not only today but also in the future.
Highly multiplexed methods represent the next step in the field of spatial proteomics and immunohistochemistry.
Why did you decide to offer CODEX multiplex imaging services and what insights does it provide your users?
We have been working with imaging for many years as part of the Human Protein Atlas (HPA) project, where we’ve used immunofluorescence and confocal microscopy to map the subcellular proteome in a variety of cell lines. This has been done in a high throughput manner, although we have been limited to look at one protein at the time. The highly multiplexed methods now available make it possible to detect many more proteins in the very same sample, and these methods represent the next step in the field of spatial proteomics and immunohistochemistry. In 2018, the facility, as part of Emma´s lab, entered the CODEX Early Access Program as one out of ten labs around the world and by this we made our entry into the field of highly multiplexed imaging. To offer CODEX as a facility service was always part of the plan.
Although there are now several platforms for highly multiplexed imaging available, we felt familiar with the type of data that the CODEX platform generates. Having the experience in microscopy and in interpreting and analyzing fluorescence images makes it easier to do proper quality control of what we see and also to establish and optimize protocols relating to CODEX. Since we were working a lot with confocal microscopy before CODEX, we are also quite picky when it comes to the resolution and the quality of the images.
Our work with the HPA has also given us extremely valuable experience in antibody validation. Even a cross-reactive antibody can generate images that looks perfect with a high specific signal and minimal background. Any antibody thus has to be carefully evaluated in the right context before trusting it and including it in a panel. Since we have been early testers of CODEX, we have seen how it developed over recent years.
To be in the position where we can now offer this as a service is very encouraging. We see that it has a lot of potential and we have several users already that have been very excited about the data that we have generated in their projects. I know that the first ones are writing their manuscripts right now and it’s really great to see how we can help disseminate this technology to the users and help them answer their biological questions not being answered before.
And we are also excited, of course, for our own research and the new antibody panels being developed in Emma’s lab.
This combination of spatial profiling and multi-omics will be needed to unravel many biological processes and mechanisms that are sometimes hard to understand.
What kinds of CODEX projects have you completed for users?
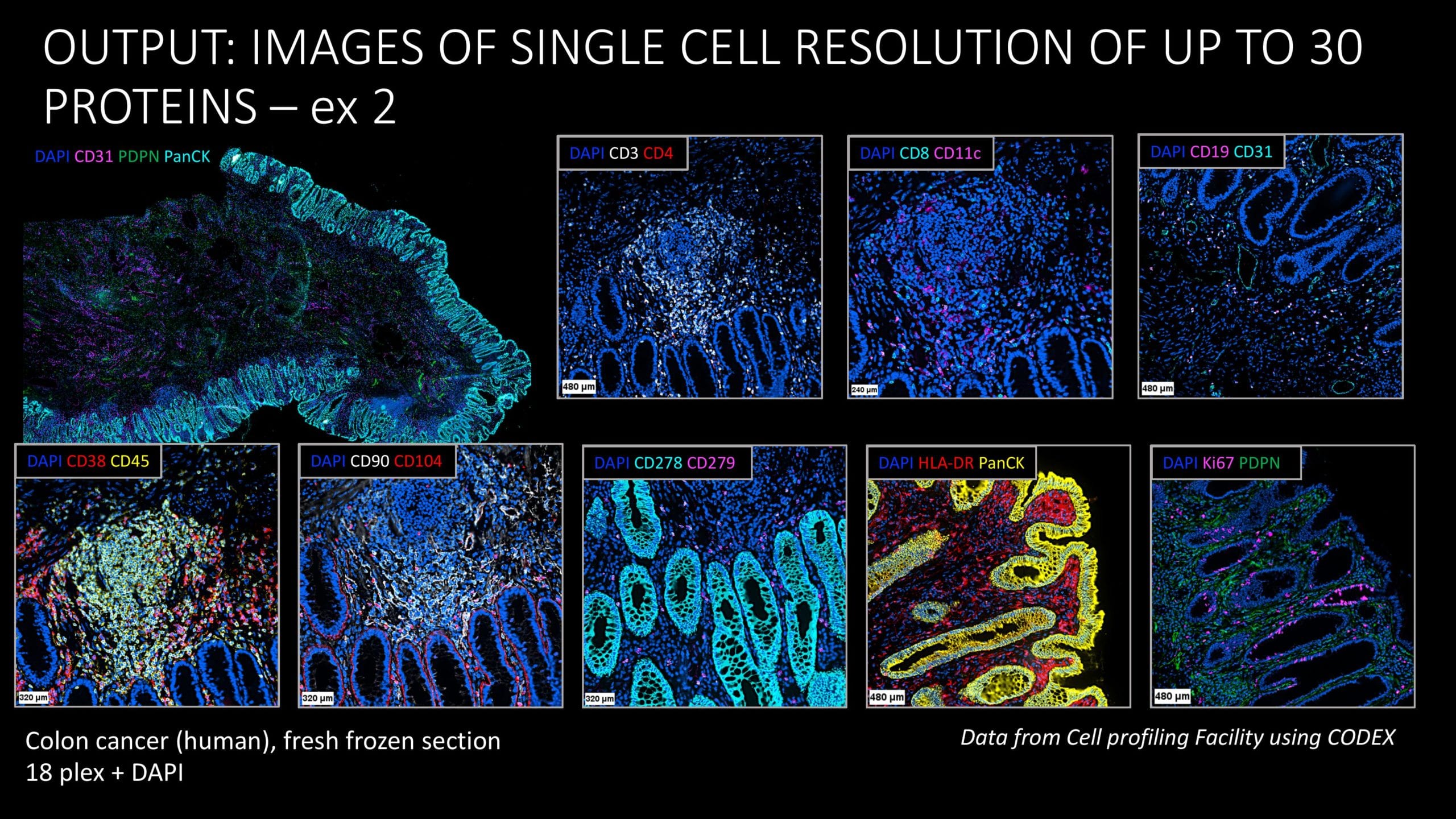
CODEX is a rather new technology. As far as I know, we are the only lab in Sweden and one of a few in Europe that is providing access to the CODEX system. This means that we have the benefit of working with many different kinds of samples. We have done projects with both fresh frozen tissues, FFPE and some tissue microarrays, including lung cancer TMAs. We have also worked with colon samples and on neuroblastoma and we will soon run a TMA of post-mortem COVID-19 samples from different tissue origin. We have also done a project on liver sections, and we’ve done some attempts with liver spheroids as well.
For the users, we provide commercially available conjugated antibodies. But we also ask the users for potential targets that they are interested in including that are not available so far. We gather this information and once we have targets that are being requested by several users, this acts as our priority list within our lab to try to conjugate these. Since we are working a lot with subcellular distribution of proteins, Emma’s lab has also generated an organelle panel of antibodies for CODEX that will soon be available for use in the facility.
We have many pioneers in the field of spatial biology at SciLIfeLab and there is another facility, in situ sequencing (ISS), that is providing service for in-situ sequencing which allows for cellular profiling of tissue sections from sequencing up to a few hundred targeted transcripts at single cell level. Another facility offers spatial transcriptomics (global mRNA profiling) with the 10x Visium technology. Our intention is to work closely with these other spatial-omics facilities at SciLifeLab and for the future try to integrate the layers of omics data we can generate.

What are the advantages of a multi-omics approach?
In Emma’s lab we also work a lot with transcriptomics data and we have seen how important it can be to gather information not only at the transcript or protein level but to integrate such omics data to better understand cellular behavior and processes. We have seen that there are proteins that are regulated at the post-transcriptional level.
The combination of genomics, transcriptomics and proteome data is already being used, but not with spatial technologies in situ. This combination of spatial profiling and multi-omics will be needed to unravel many biological processes and mechanisms that are sometimes hard to understand.
Our aim is to make sure that we deliver data that we believe in.
What happens with the imaging data you collect from CODEX?
We work with the CODEX Multiplex Analysis Viewer (MAV). What we always do is a quality control of the data generated. For that, we use MAV to explore and look at all of the intensities for every marker and to see whether the staining pattern makes sense. And then we can do some sort of basic analysis, again focusing on the quality control. Our responsibility is, first of all, to make sure that the run itself has been working as it should, and that we believe in the antibodies used and that the staining makes sense.
But if the users have very specific questions that they define when they submit the project application, then we can look more into that once we are doing the QC. Since analysis can take a lot of time, we are not providing too much service on that right now. Our aim is to make sure that we deliver data that we believe in. Then it’s up to the user to really find the biological answers from that.
We are trying to get more hands so that we can take on a bigger part of analysis. But for now, we have a follow-up meeting with the users where we give them an introduction for how they can work more with MAV to do their analysis. Our goal – although with limited resources – is to give the user some concrete results from their data, to help them bring their project forward.
How can researchers get in touch with you to initiate a project?
We are trying to get more hands so that we can take on a bigger part of analysis. But for now, we have a follow-up meeting with the users where we give them an introduction for how they can work more with MAV to do their analysis. Our goal – although with limited resources – is to give the user some concrete results from their data, to help them bring their project forward.
We have a page for the Cell Profiling Facility on the SciLifeLab website. But the best way to reach us is to send an email to cellprofacility@scilifelab.se. We can provide further instructions from there.