We were in Palm Springs last week for the Association of Biomolecular Resource Facilities (ABRF) 2020 Annual Meeting. Core lab professionals came together to learn about cutting-edge advances that they can bring to their facilities.
Some of this year’s talks revolved around multi-omics, single-cell techniques, and spatial analysis. This reflects scientists’ growing need for analysis tools that can provide multi-dimensional, spatial information at high resolution.
We showcased our CODEX® and Phenoptics™ platforms at our booth in the exhibit hall and during an evening workshop.
Cell Sciences Imaging Facility (CSIF) at Stanford Implements Single-Cell Spatial Tissue Profiling
At our workshop, we were joined by Jon Mulholland, Director of the Cell Sciences Imaging Facility at Stanford University, who shared his experience implementing CODEX as a shared resource.
About CSIF
The CSIF is organized into three interdependent cores which provide access to state-of-the-art imaging technologies and analysis software for multidimensional analysis of cells and tissues. The Fluorescence Microscopy Core provides services like super-resolution microscopy and multi-photon imaging, while the Electron Microscopy Core offers services such as correlative light to electron microscopy and high-resolution field emission scanning electron microscopy.
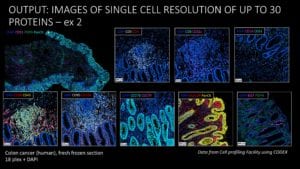
CODEX was installed as part of the Multiplexing and Array Tomography Core, which provides multi-marker, 3D volumetric reconstruction using Array Tomography and highly multiplexed, single-cell, spatial tissue analysis using the CODEX platform.
Identifying the need
Jon covered the steps involved in introducing new instrumentation and starting a new service in his core facility. First, he and the CSIF team identify a need from their user group and evaluate how often and when this new service or instrument will be required. They justify the need by trying to answer key questions such as: Is the instrument or service in question the only way to provide a type of data? Will this instrument or service advance the field significantly faster and improve data quality?
Budgeting and Resources
If the answers are yes, they move on to creating a budget for the next five years, identifying operating costs, a plan to recoup those costs, and if any subsidies will be offered. They also determine a physical space in the lab for the instrument. Finally, CSIF acquires funding for the instrument and determines if an existing staff member will provide the service, or if someone must be hired. The service can then begin.
The advantages of CODEX
In the case of CODEX, CSIF users had a need for quantitative, highly multiplexed, multiparametric, spatial data. They were looking for efficient labeling of many molecules in a tissue with little to no spectral overlap and high dynamic range. In terms of visualization and analysis, they needed tools that could map reporters to tissue areas, segment cells and determine cell types and functional states, while maintaining their environmental context.
Traditional immunofluorescence methods didn’t meet these needs. Only a limited number of biomarkers (~3-6) can be applied due to spectral overlap limitations. Mulholland’s group also considered imaging mass cytometry (IMC) and multiplexed ion beam imaging (MIBI). However, both technologies use rare-earth metal isotopes for tagging, of which there are only ~32 available. They also require specific, expensive reagents and equipment, and tissues analyzed with these methods cannot be reused for downstream analysis.
Instead, CODEX was chosen to meet this need due to multiple advantages. CODEX is flexible – any antibody can be used regardless of the host species, and labeling the tissue with antibodies is done in a single step. The oligo denaturing step in the CODEX workflow is gentle to the tissue, allowing for the sample to be reused. Since CODEX is an automated platform, you can set up your experiment and walk away. Jon highlighted that the CODEX technology is cost-effective compared to alternatives, and since it was invented in the Nolan lab at Stanford, CSIF had local expertise available as well.
Antibody validation at CSIF
Using CODEX, CSIF performs tissue profiling using Akoya’s inventoried and screened antibodies, panels validated by Akoya (or other researchers), and custom validated antibody conjugates and panels.
Staining in a tonsil tissue microarray (TMA). Image courtesy of Jon Mulholland.
Jon discussed some techniques his lab uses for custom antibody validation. He suggested selecting antibodies that have been previously used for immunofluorescence and screening more than one potential antibody to the same protein, if possible. He emphasized the importance of validating antibody specificity on well characterized tissue with defined structures and comparing the localization in normal samples to diseased tissue samples (using tissue microarrays, if possible). CSIF evaluates staining for subcellular localization, cell type and tissue localization, signal intensity, signal to noise ratio, and antibody titration. More information on custom conjugating your own antibodies can be found here.
The CSIF has had success installing CODEX in their facility to provide highly multiplexed, spatial tissue imaging to their users. CSIF hired a new staff scientist to lead the CODEX pipeline development and services and received training from Akoya. They currently provide services to both Stanford research groups and external researchers, including industry scientists.
We’d like to thank Jon for sharing his experience and speaking at our ABRF workshop. Click here to learn more about CODEX for spatially resolved, highly multiplexed tissue biomarker analysis.