This post is part of a two-part series on Akoya’s software tools for spatial analysis. Part 1 covers the CODEX Multiplex Analysis Viewer.
As researchers attempt to characterize cellular activity within the tissue microenvironment, it’s critical for them to have the tools they need to extract meaningful biological information from tissues. This is essential for basic research and drug development studies, as well as for analyzing the tumor microenvironment to determine cancer subtypes, validate drug targets, and stratify patients in translational oncology.
The Phenoptics™ solution includes a range of biomarker discovery tools which combine the latest technologies with trainable, automated algorithms and easy-to-use interfaces, enabling you to extract new, exciting discoveries from your data.
- Annotate — Phenochart is a whole slide contextual viewer that enables viewing and annotation of Brightfield and Fluorescent digital slides scanned on the Vectra 3 and Vectra Polaris
- Analyze — inForm® Tissue Analysis Software is an automated tissue analysis software package enabling comparative studies of multiple markers and specimens
- Report — phenoptrReports provides powerful, easy-to-use tools to analyze spatial relationships between cellular phenotypes and visualize their relationships on the tissue, displaying results in shareable, easy-to-read data reports
In this post, we’ll discuss how inForm enables researchers to visualize, analyze, quantify, and phenotype cells with multiplexed immunofluorescent stained FFPE tissue sections.
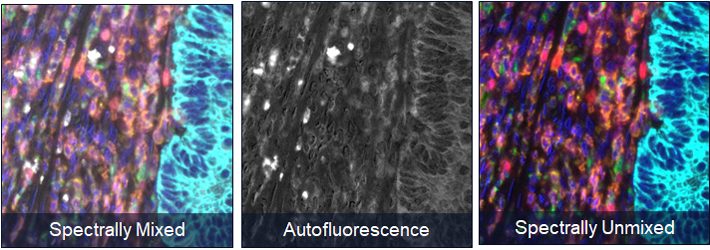
The power of spectral unmixing
One of the key features of the Phenoptics solution is multispectral imaging. By imaging tissues with our Vectra and Mantra systems it becomes possible to capture cellular interactions occurring between cells across an entire tissue section.
Once you’ve captured a tissue image, you can annotate hotspots of immune activity in Phenochart and then move to inForm for further analysis, including spectral unmixing. inForm separates the signals from the fluorescent reagents placed on the tissue into invidiual bins. By first isolating them, we can quantify all the signals present on a tissue, regardless of intensity or signal overlap.
inForm also isolates autofluorescence into its own channel. This ensures that you are accurately quantifying the immune interactions occurring in the tissue without false-positive signal from background signal. inForm is the only commercially available tissue analysis software with this powerful spectral unmixing engine.
Tissue and cell segmentation
After spectral unmixing, the next step is to segment your tissue types based on tissue morphologies, dividing the image into specific areas that correspond to specific types of tissue (e.g. cancer, necrosis, stroma, etc.) or other structures of interest. This is accomplished using inForm’s easy-to-train, AI-powered, feature recognition algorithms. After drawing only a few representative training regions, the tissue segmenter can be trained to automatically locate similar regions of tissue in each image.

Once tissues are segmented, users can identify all the individual cell types. inForm’s “Adaptive Cell Segmentation” uses an adaptive thresholding algorithm to identify nuclear pixels. Densely packed and overlapping nuclei can be easily identified using nuclear assist markers regardless of staining heterogenerity. Segmenting cells into their respective compartments (nuclear, cytoplasm, membrane), enables the user to extract and analyze fluorescence data on a per-cell or per-compartment basis.
Following segmentation, each cell is provided with a unique cell ID, both within the field and on the whole slide (if you’re imaging with MOTiF™ on the Vectra Polaris). Cells are also given x and y coordinates, so that once you’ve assigned them a phenotype, you can begin to perform spatial analysis to study their interactions.
Phenotype cells with inForm
inForm enables the automated classification of cell phenotypes using a trainable, machine-learning algorithm. Using only a small training set of user-assigned phenotypes, inForm can quickly identify remaining cells and provide per-cell confidence statistics. Users can verify phenotypic accuracy and quickly update the training set to ensure minimal error in cellular classification across a tissue section.

Phenotyping labels cells with different colored dots within the boundaries identified by cell segmentation. Each dot is representative of a different phenotype within the tissue (i.e. PD-L1+, panCK+, etc.). Users can also add phenotypes representing cells where multiple biomarkers are expressed (i.e. PD-L1+/panCK+ cells). Since fluorescent signals are spectrally isolated, researchers can confidently identify phenotypes arising from low expressing epitopes and count low phenotypic expressors.
Automate analysis of inForm outputs with phenoptrReports
inForm Analysis generates a large amount of data. This may leave users at a loss as to where to begin to understand the biology of their tissue. phenoptrReports helps to address this challenge, displaying results in clear, shareable data reports.
Data is made actionable through powerful analytics tools which enable users to study tissue microarrays (TMAs) and regions of interest and perform advanced phenotypic reporting — an easy way to identify multipositive cells for expression and lineage markers.
Phenotypic spatial maps allow you to look at single-positivity and multipositivity phenotypes directly on the tissue and understand how they’re interacting through nearest neighbor and proximity analyses.
The reports generated by phenoptrReports are customizable, shareable, and easy-to-read. Audit trails of the user’s actions are generated so those same analyses can be repeated by other researchers.







